Scaffold Hopping User Guide
1: Introduction to the Algorithm
Scaffold Hopping module can offer you a rapid generation of novel lead molecules with similar or more potent bio-activities, but improved physicochemical properties, ADME properties, toxicities, selectivities and so on.
Along with lead optimization, Scaffold Hopping module can also help arouse ideas for novel chemical series to a known drug.
2: Start to Use
2.1 Upload molecule and get 3D conformer
iDrug provides two methods of inputting data, including inputting SMILES, and uploading SDF file with 3D conformation.
-
Input SMILES
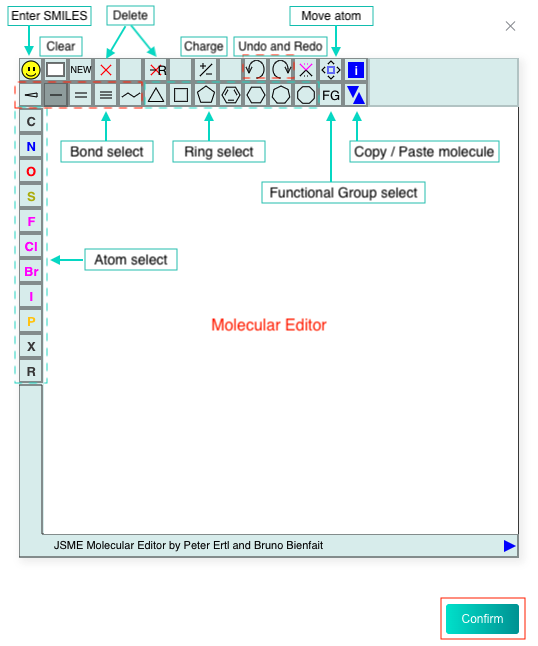
Select the "Input your SMILES" check box, then enter one SMILES in the text box, or click the "DRAW" icon at the end of the text box to pop up the molecular editor, in which you can draw the molecular structure and click "Confirm" to convert it to SMILES which will be displayed in the text box.
Click the "Get 3D conformation" button, then iDrug will generate 10 3D conformers for this molecule, which will be displayed below. After selecting a suitable 3D conformer as the reference conformation of this molecule, you can click the "Next" button to enter the next step.
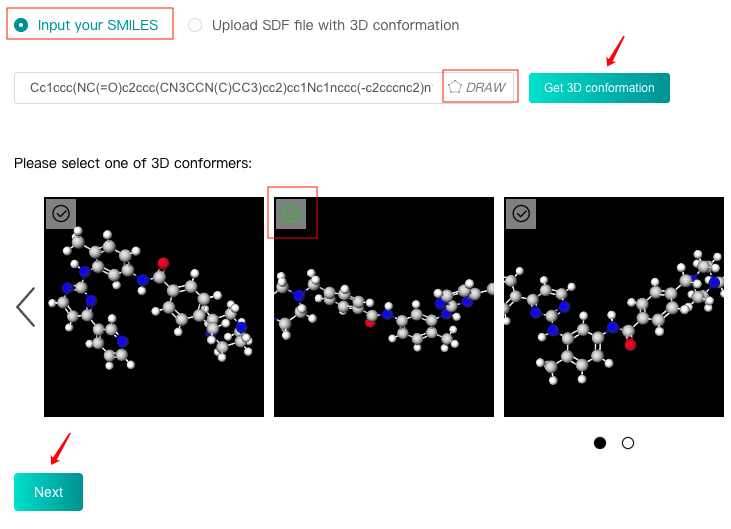
-
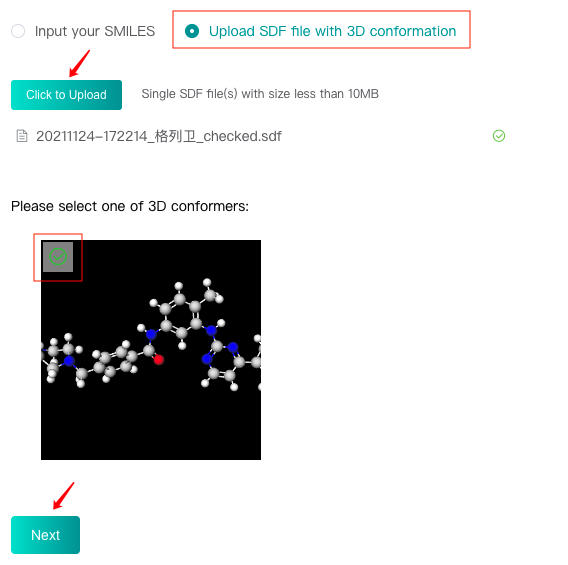
Upload SDF file with 3D conformation
Select the "Upload SDF file with 3D conformation" check box, then click the "Click to upload" button to upload an SDF file, and click "Get 3D conformation" button to display the conformer in this file below. Select this conformer and then click the "Next" button to enter the next step.
There can only be one molecular conformer in the uploaded SDF file, which must be 3D conformation, and do not support protonation.
2.2 Input scaffold of molecule
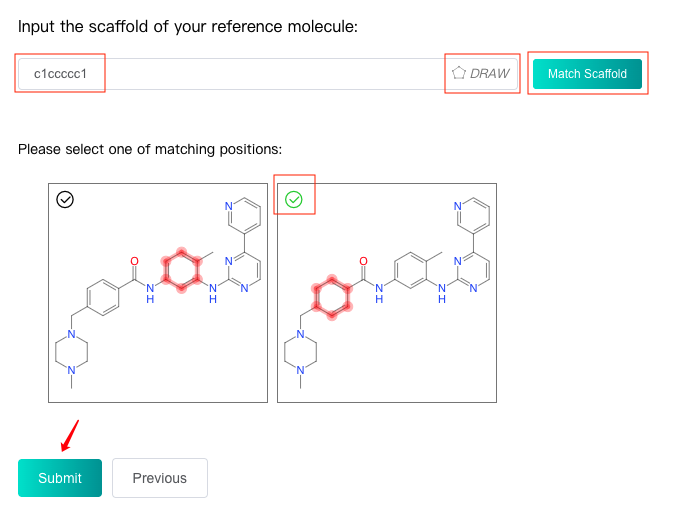
Enter the SMILES of scaffold which will be replaced in the reference molecule in the text box, or click the "DRAW" icon at the end of the text box to pop up the molecular editor, in which you can draw the scaffold structure and click "Confirm" to convert it to SMILES which will be displayed in the text box.
Click the "Match Scaffold" button, iDrug will match the scaffold in the reference molecule and display the matching positions below. Select one of the matching positions and then click the "Submit" button to submit task and wait for the task to be completed.
If the scaffold cannot be matched successfully in the reference molecule, an error will be reported which means users should check the uploaded molecule or scaffold. If multiple positions are matched, only one matching position can be selected for replacement.
2.3 Submit task
After completing the above steps, click the "Submit" button to submit the task. If "Your task has been created successfully!" is displayed, the submission is successful.
If the task is completed successfully, the user's registered mailbox will receive the notification of task completion. Usually, the calculation time of a task takes several hours.
In order to allocate the limited resources reasonably, we limit the user's task quota. Hover the mouse over the "Submit" button or the "Next" button to see the restriction and usage. The "Submit" button and the "Next" button will be disabled when quota is insufficient.
3. Query the Result of Your Task
3.1 Query history
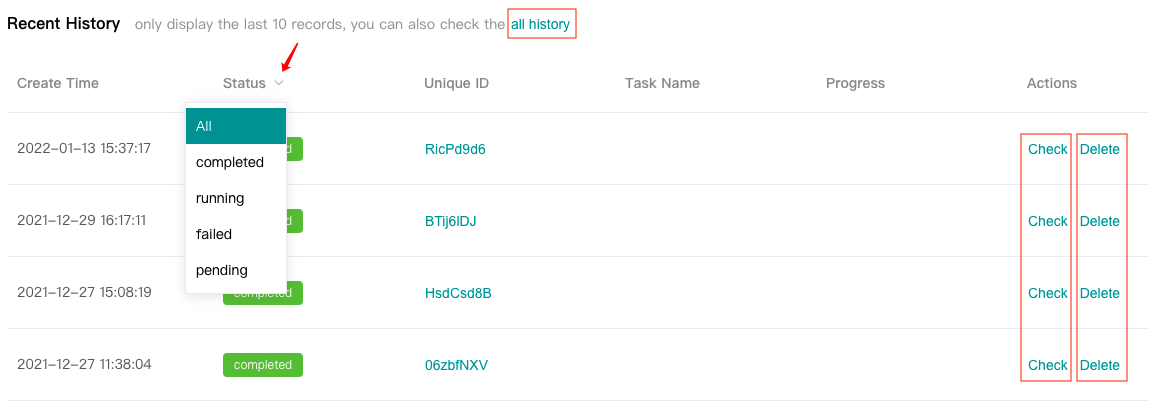
Users can view the "Recent History" below, which only displays the last 10 history records, or click the "all history" to see all history records.
Users can "Check" completed tasks (marked in green).
Users can "Delete" completed or failed or pending tasks. Tasks in running status can not be deleted.
Click the drop-down arrow next to "Status" to filter according to the task running status.
3.2 Query results
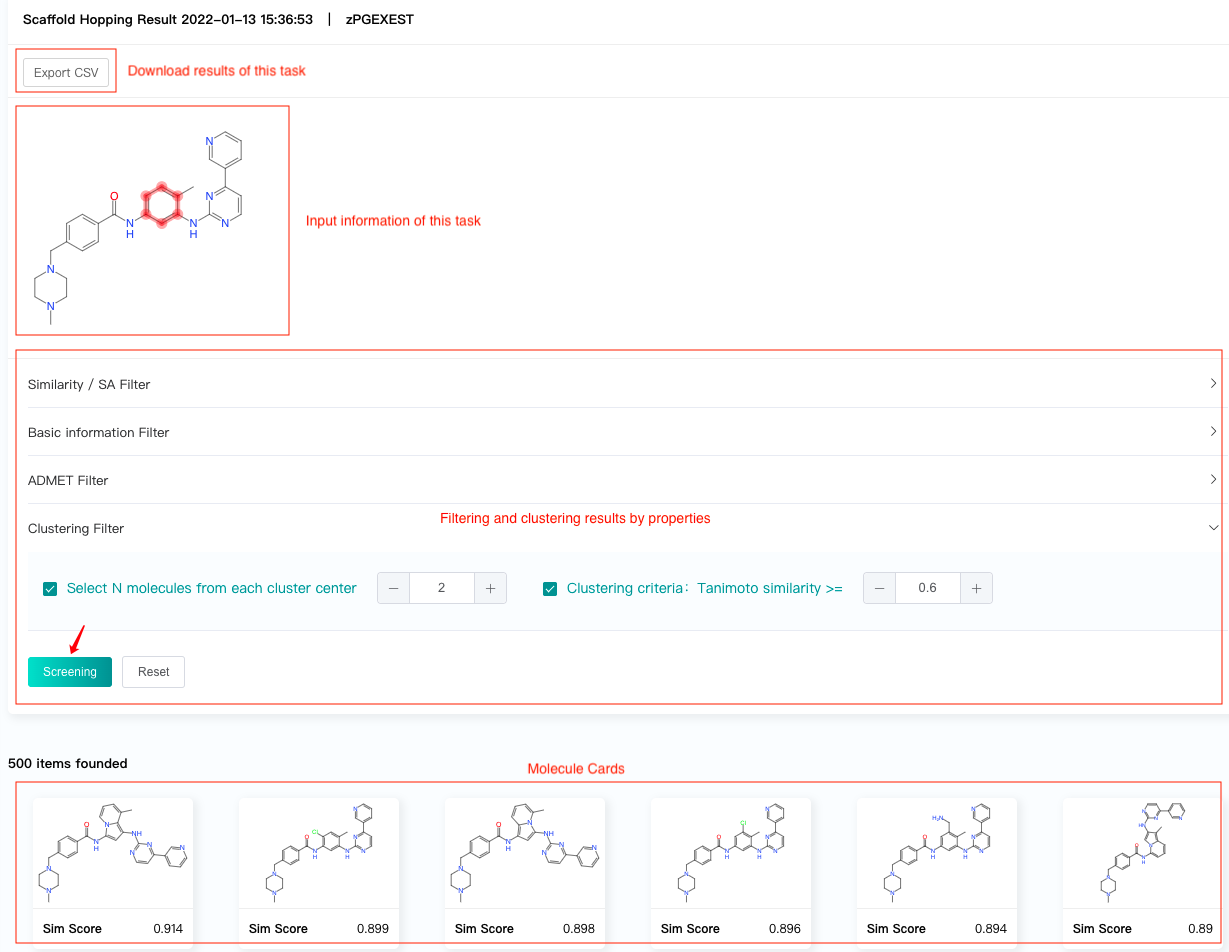
Click the "Check" button on the history records to view the task results.
The result interface displays:
-
Matching position of the input molecule and scaffold, which help users to confirm the input information of the task.
-
Filtering and clustering results by properties.
-
Molecule cards, which show the predicted similarity scores of the generated molecules.
-
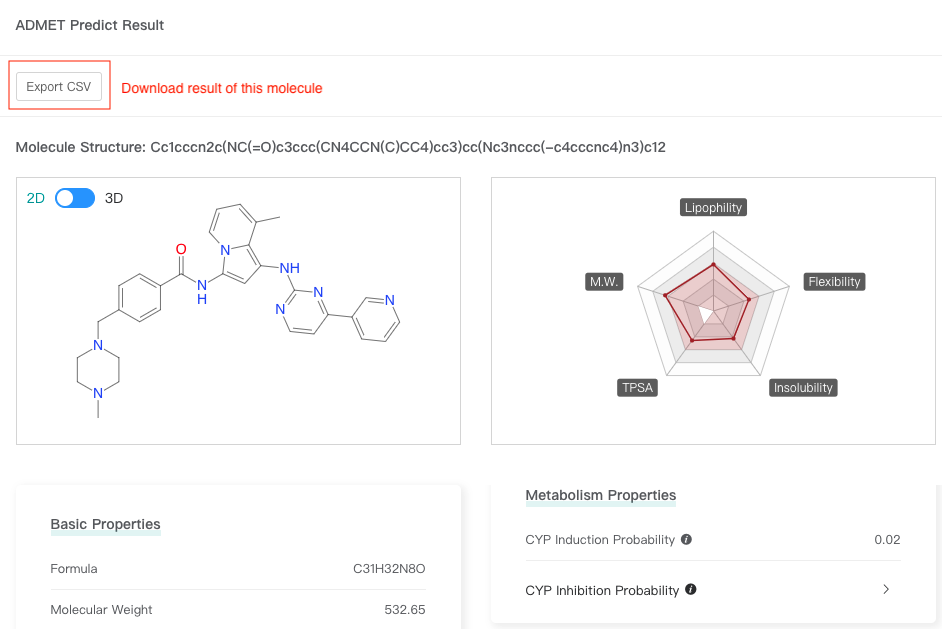
Click one molecule card to view the molecular attributes and ADMET attributes (predicted values) of this molecule.
3.3 Download results
In the result interface, click "Export CSV" and select the attributes to be retained to download all results.
Click one molecule card to view the attributes of this molecule, and then click "Export CSV" to download result of this molecule.