Molecule Generate User Guide
1. Introduction to the algorithm
Generative chemistry is a very important for de-novo drug design, which can find new chemical scaffolds beyond existing screening libraries. The molecular generation algorithm of Tencent is to learn the structure information of small molecules related to protein targets in a known chemical space. Currently, we support 319 kinase and 52 GPCR targets to generative chemistry.
During this process, our algorithm can sample molecules from the projection of the molecular space towards different targets and then generate novel molecules with activities. Or you can select a specific target and upload a reference compound. Our algorithm can also generate novel molecules by changing motifs of the reference compound, with maintaining bioactivity.
2. Start to Use
Two ways are supproted by our Generative chemistry model: only target or target with ONE reference compound.
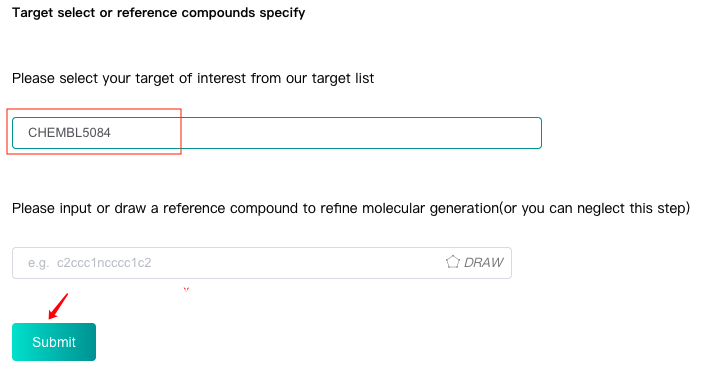
2.1 Input target of interest only
Please select or type in one target that you are interested in from our target list and click the "Submit" button to submit the task. Then our model will generate one batch of molecules with their basic and predicted ADMET properties based on molecules of this target.
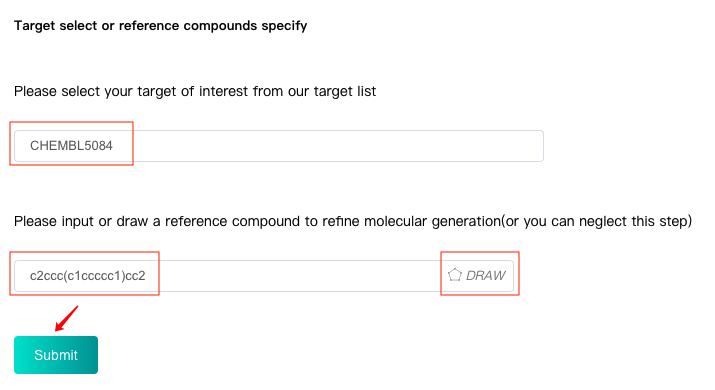
2.2 target with one reference compound
Firstly select or input your target.
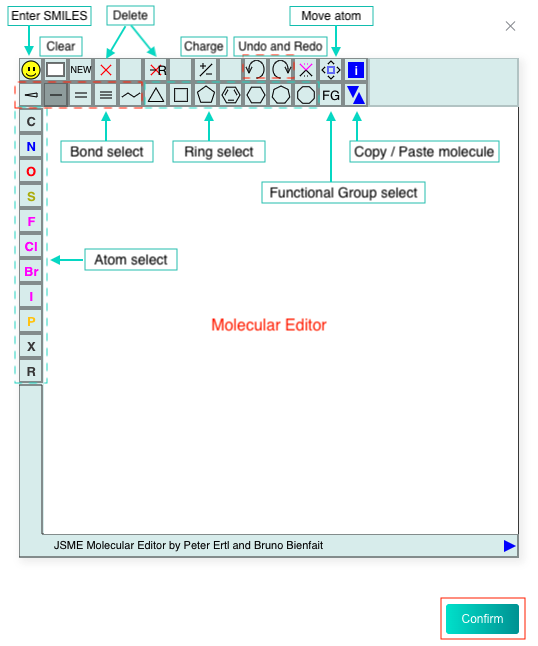
Then input the SMILES of reference compound in the text box or or click the "DRAW" icon at the end of the text box to pop up the molecular editor, in which you can draw the scaffold structure and click "Confirm" to convert it to SMILES which will be displayed in the text box.
Finally click the "Submit" button to submit. By this way, the algorithm will refine the molecular generation process by the reference compound and generate a batch of novel molecules with basic and predicted ADMET properties.
2.3 Submit task
Click the "Submit" button to submit the task. If "Your task has been created successfully!" is displayed, the submission is successful.
It usually takes 40 minutes or more for a task, depending on the complexity of the input molecule. Please be patient.
In order to allocate the limited resources reasonably, we limit the user's task quota. Hover the mouse over the "Submit" button to see the restriction and usage. The "Submit" button will be disabled when quota is insufficient.
3. Query the Result of Your Task
3.1 Query history
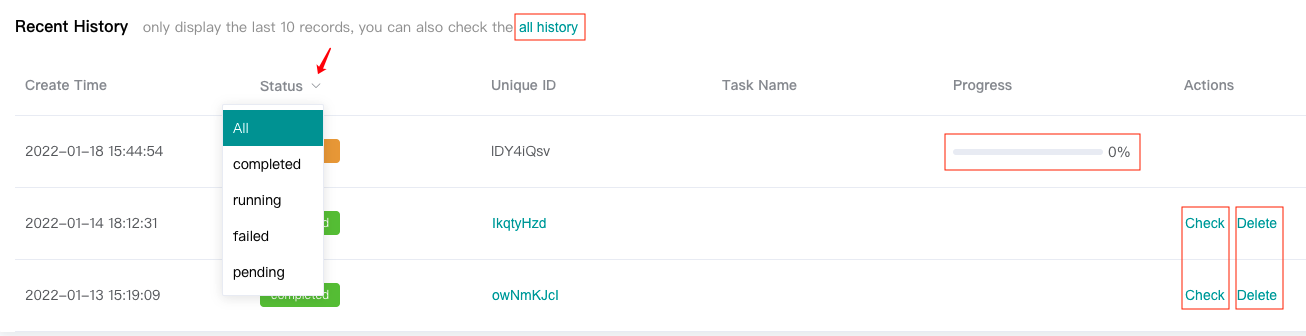
Users can view the "Recent History" below, which only displays the last 10 history records, or click the "all history" to see all history records.
Users can view the task progress of running task through progress bar.
Users can "Check" completed tasks (marked in green).
Users can "Delete" completed or failed or pending tasks. Tasks in running status can not be deleted.
Click the drop-down arrow next to "Status" to filter according to the task running status.
3.2 Query results
Click the "Check" button on the history records to view the task results.
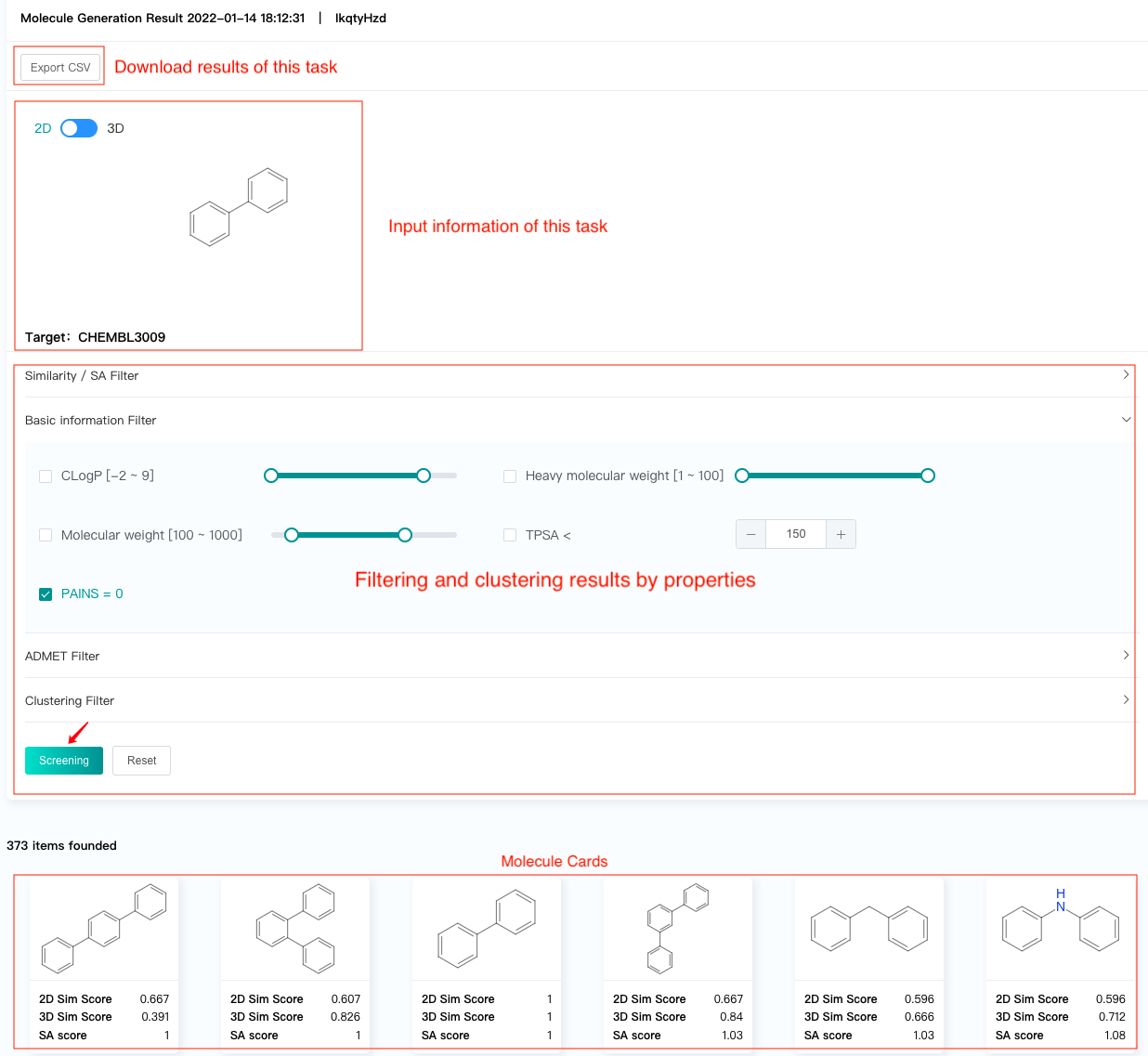
The result interface displays:
-
Target ID of the task and the 2D/3D conformer of the reference molecule (if any) to confirm the input information of the task.
-
Filtering and clustering results by properties.
-
Molecule cards, which show the predicted 2d & 3d similarity scores and SA scores of the generated molecules.
-
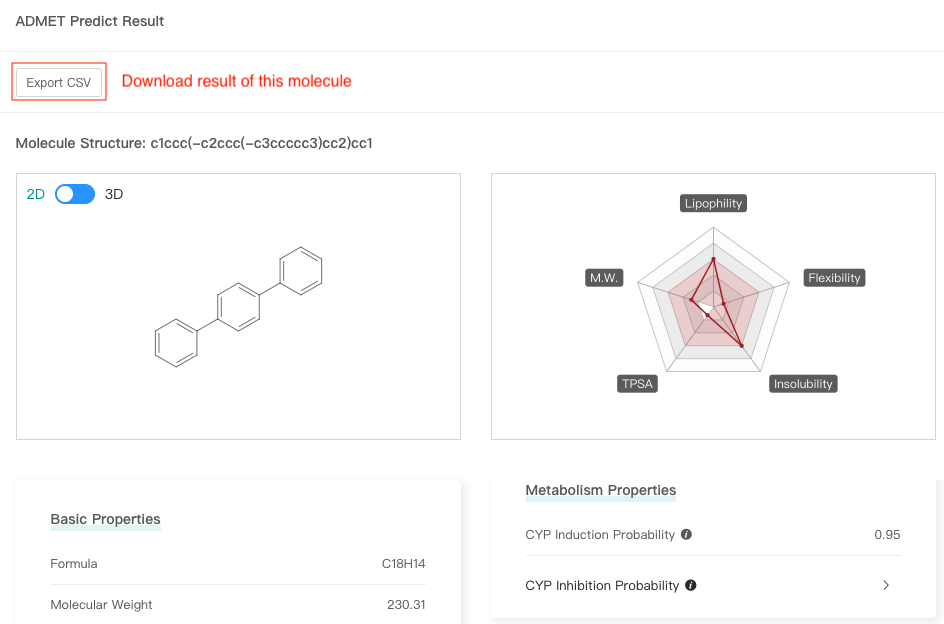
Click one molecule card to view the molecular attributes and ADMET attributes (predicted values) of this molecule.
3.3 Download results
In the result interface, click "Export CSV" and select the attributes to be retained to download all results.
Click one molecule card to view the attributes of this molecule, and then click "Export CSV" to download result of this molecule.